Genomic Evaluations
Genotyping and genomic evaluations have transformed the mating, marketing and selection of male and female dairy animals as parents of the next generation.
Genomic Selection Fuels Genetic Gain
Genomic technology has resulted in tremendous genetic gains, more accurate and earlier prediction of animals’ genetic merit and changes in herd culling and breeding strategies. Use of genomics also expedites evaluations for new traits when phenotypes are limited.
Genomic selection refers to decisions based on genomic breeding values. Genotypes, pedigree information and recorded phenotypes are combined to predict animals’ genetic merit.
Reliable genomic evaluations depend on a continuous flow of accurate data – both genotypic and phenotypic – to establish and refresh the reference population, measure performance, document genotypes and calibrate genomic results. The National Cooperator Database is the foundation of both traditional genetic and genomic evaluations.
female phenotypic records
dairy animal genotypes
genotypes added per year
CDCB Genomic Evaluations
In the U.S. genomic evaluations are expressed as GPTAs, or Genomic Predicted Transmitting Ability – similar to the traditional evaluations that include phenotypic results. CDCB releases genomic evaluations weekly, monthly and triannually, dependent on an animal’s status. Evaluations of newly-genotyped animals are released weekly for immediate decisions around mating, culling and herd management
Genomic Data Flow into Database
Genomic evaluations begin with the decision to genotype a dairy bull, cow or heifer. Biological (DNA) samples and data flow through a proven process that results in CDCB genomic evaluations.
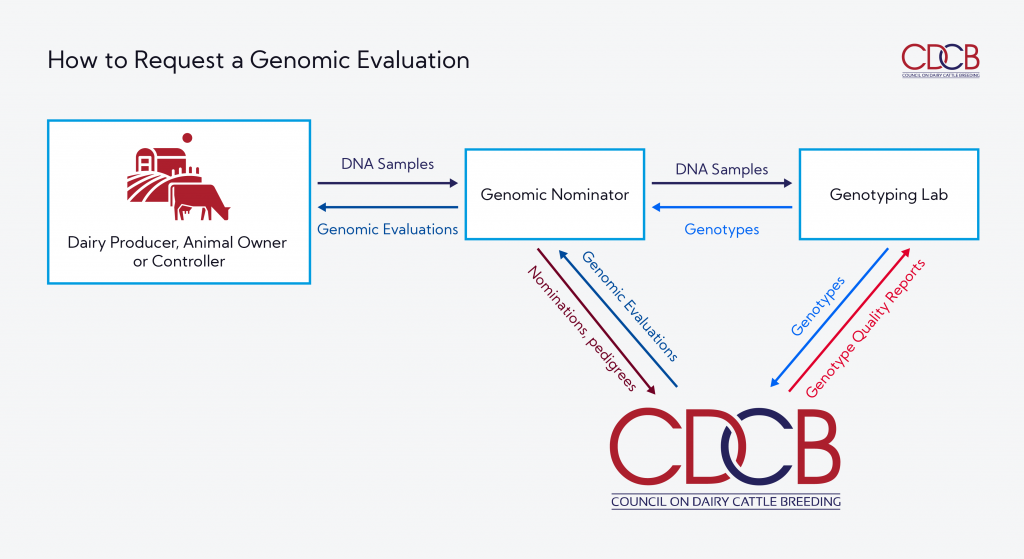
CDCB receives pedigree and other information from genomic nominators and genotype data from certified laboratories. Validated and accurate data are then included in the genomic evaluations, following CDCB rules of publication.
Nomination
CDCB-certified genomic nominators – like breed associations, artificial insemination (A.I.) companies and genotyping labs – are a critical link. Genomic Nominators are responsible to:
- Collect DNA samples from the animal owner
- Transfer DNA samples to a certified genotyping lab
- Provide pedigree and other animal information to CDCB
- Distribute CDCB results to the animal owner or marketing organization
- Collect initial fees and forward to CDCB
- Meet CDCB certification requirements
Nominators provide critical information for the CDCB national cooperator database.
- Animal pedigree, important for the evaluation and parentage validation
- Association between animal ID and genotype sample ID
- Fee code, each animal is assigned a fee for the service provided.
- Type of service. Most genotypes are for genomic evaluation results, and some are only for parentage check.
Certification
To maintain quality results, nominators and genotyping labs must be certified before submitting data to the CDCB national cooperator database. Their performance is monitored routinely and audited annually.
Genotyping
CDCB accepts genotypes from certified laboratories and international evaluation centers. Genotyping Labs are responsible to:
- Extract DNA from samples
- Prepare single-nucleotide polymorphism (SNP) genotypes
- Provide summary information back to the genomic nominator
- Transfer the raw genotypes to the CDCB database
- Meet CDCB quality certification standards
Most animals are genotyped with chips that have between 40,000 and 80,000 SNP markers.
Validation
The SNP genotypes submitted to CDCB are evaluated for call rate, portion heterozygous, and parent-progeny consistency. At CDCB, each animal genotype is checked against its parents and a grandparent (if the parent has not been genotyped). Each genotype is compared with all others in the CDCB database to discover identical genotypes or parent-progeny relationships not in the pedigree. These checks ensure the genotype is assigned to the correct animal.
The CDCB database can store multiple genotypes for the same animal, as it relies on chip identification and sample location on the chip (barcode and position).
Genomic Evaluations
Genotypes declared “usable” (those who pass all quality controls and have a complete nomination) are considered to compute the genomic evaluation.
First, relevant SNPs are extracted from the database. Currently, 78,964 SNPs are used in CDCB genomic evaluations. Genotypes from chips of differing SNP densities are included in the evaluation by using imputation via FindHap algorithm to fill in the SNPs that are not observed. Traditional evaluations are de-regressed to make data more like individual records, as explained in Wiggans et al. (2011). Methodology to obtain SNP effects and the process to obtain a full genomic prediction is explained in VanRaden et al (2009). Genomic evaluations are then distributed according to the CDCB evaluation schedule
For Male Animals: How to Receive a U.S. Genomic Evaluation

For Female Animals: How to Receive a U.S. Genomic Results

Rules of Publication for CDCB Genomic Evaluations
Several steps are required to obtain a genomic evaluation on a dairy animal, and guidelines apply to animals marketed using the U.S. genetic values.

Official evaluations for U.S. animals (males and females) and foreign female animals will be published with each official monthly genomic evaluation. An exception is for animals marketed using CDCB genomic evaluations (i.e., animals with A.I. service fee paid), whose evaluations are published in the triannual runs.
No animal can be marketed using U.S. evaluations until included in the NAAB cross-reference program and their A.I. service fee is paid (or waived).
Genomic evaluations of foreign bulls are provided only to the nominator and only until 15 months of age, unless an A.I. service fee has been received by CDCB.
CDCB Fee Schedule
CDCB genomic fees reward dairy producers that provide the most data or information of the greatest value to the CDCB national cooperator database. The fee schedule encourages producers to increase the data they provide to the system, as that improves evaluation accuracy. For example, there are incentives in the fee schedule for producers to submit health events to the CDCB database, supporting reliable evaluations for disease resistance traits.
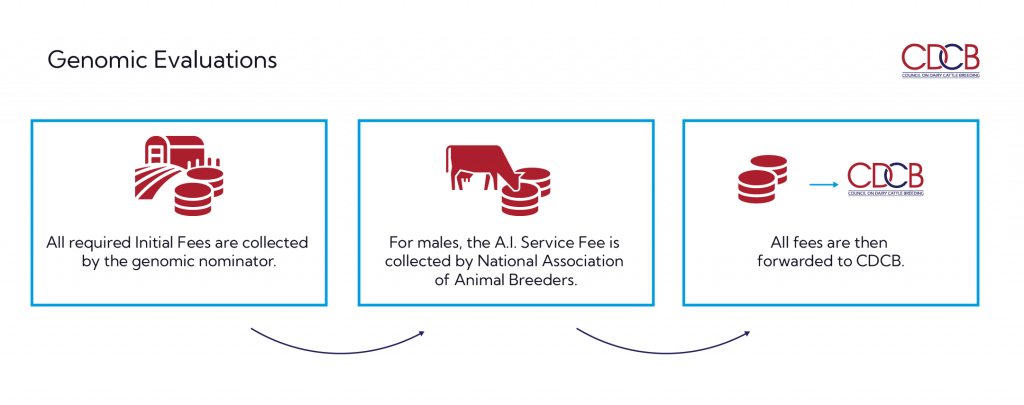
- All required Initial Fees are collected by the genomic nominator.
- For males, the A.I. Service Fee is collected by National Association of Animal Breeders.
- All fees are then forwarded to CDCB.
The female and initial male fees are charged only for the animal’s first genotype submitted for genomic evaluation.
There will be no refund of fees, except for errors generated by CDCB. Fees apply, even if the genomic test results do not work to the submitter’s satisfaction or when a male is not placed into service.